Pathway Commons
Access and discover data integrated from public pathway and interactions databases.
* Development site (current release is here)
Training
Tools
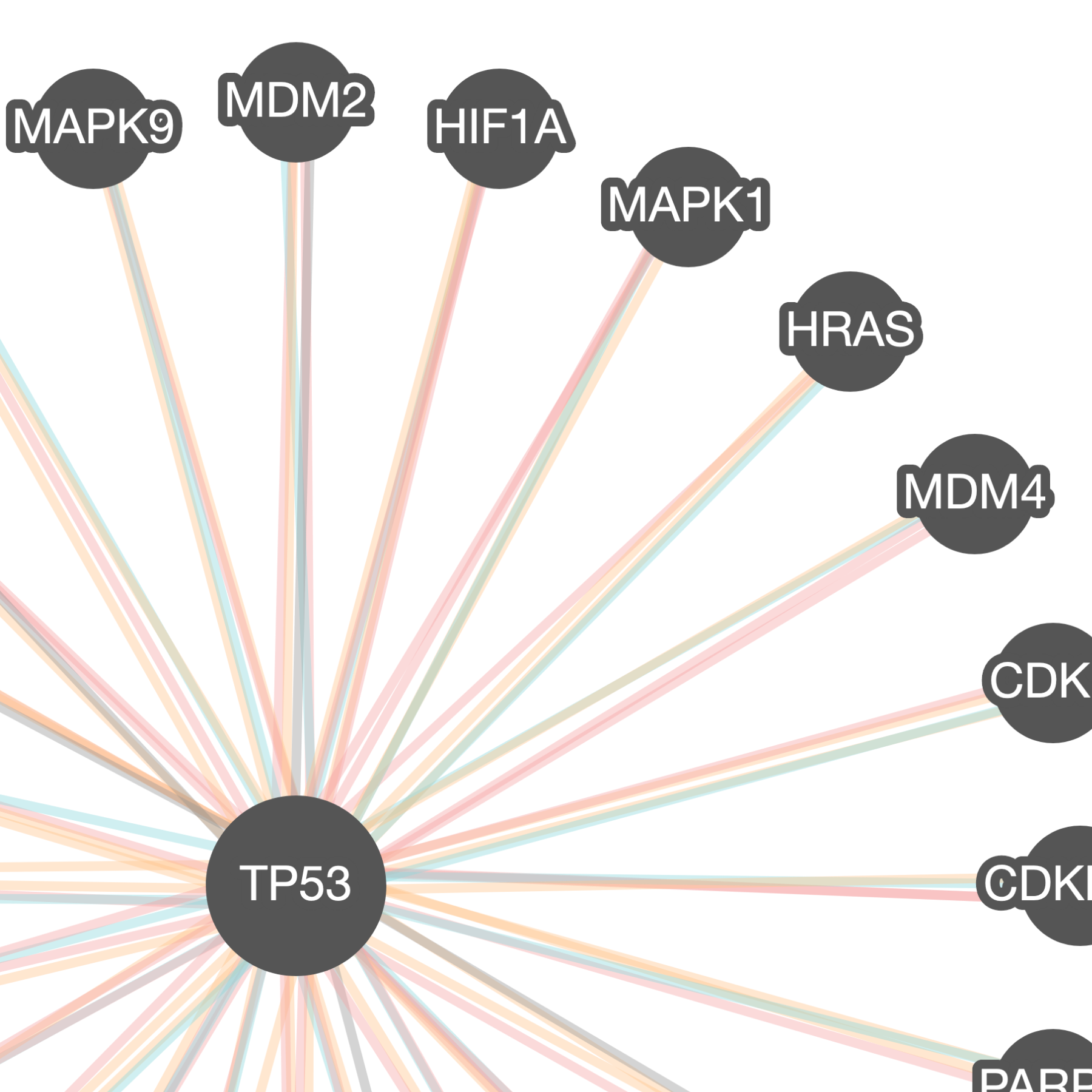
PCViz
Data visualization and analysis
Developer resources
Frequently Asked Questions
What is Pathway Commons?
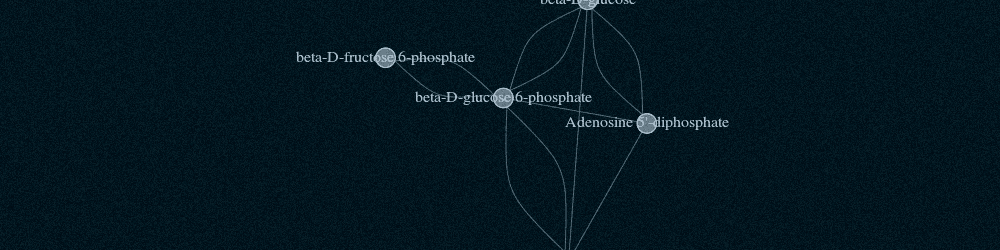
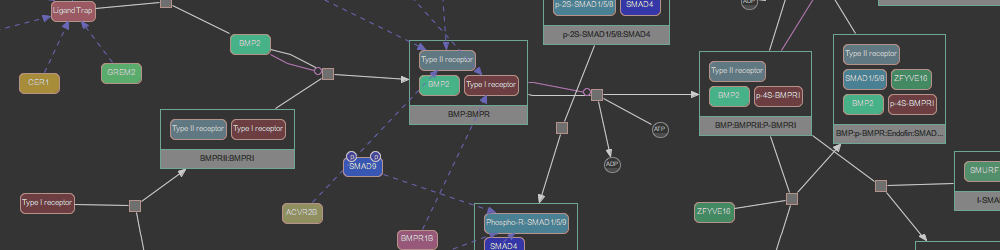
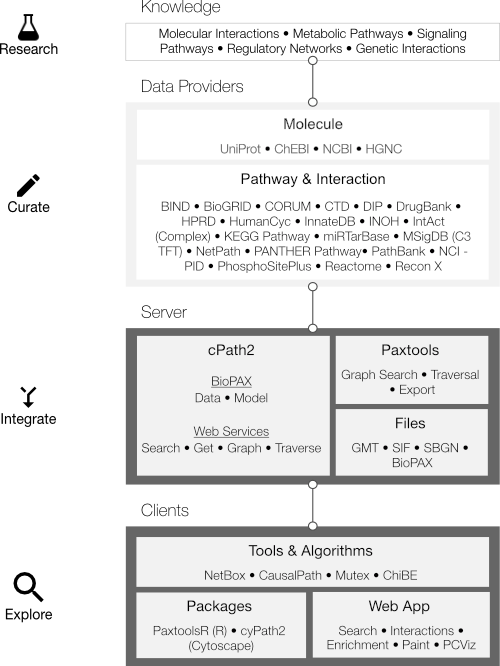
Pathway Commons aims to collect and disseminate biological pathway and interaction data. Data is collected from partner databases and is represented in the BioPAX standard. By representing data in BioPAX, Pathway Commons is able to provide a detailed representation of a variety of biological concepts including: Biochemical reactions; gene regulatory networks; and genetic interactions; transport and catalysis events; and physical interactions involving proteins, DNA, RNA and small molecules and complexes.
Is it free to use?
Yes. All of the data provided by Pathway Commons is free! In particular, Pathway Commons distributes pathway information with the intellectual property restrictions of the source database; Only databases that are freely available for academics are included. All of the software that we provide is open-source.
What can I do with this information?
- Researchers
- A common practice is to identify pathways that are enriched in gene expression data. To this end, Pathway Commons provides gene set database file downloads for direct use in Gene Set Enrichement Analysis (GSEA).
- Softare developers
- Download and incorporate biological pathway data as part of metabolic and gene pathway analysis software in BioPAX Level 3 format. Details about the BioPAX format
How can I access the data?
- Web
- Search and view pathways and interactions
- Desktop software
- Retrieve, view and edit Pathway Commons data using the Chisio BioPAX Editor (ChiBE)
- Search and analyze Pathway Commons data from Cytoscape
- Search and analyze Pathway Commons data using the R programming language
- Programmatically
- Search, retrieve and navigate over Pathway Commons data using our set of web services
- Use semantic queries for Pathway Commons data at our SPARQL endpoint
Does Pathway Commons compete with other pathway databases?
Pathway Commons does not compete with or duplicate efforts of pathway databases or software tool providers. Pathway Commons will add value to these existing efforts by providing a shared resource for publishing, distributing, querying, and analyzing pathway information. Existing database groups will provide pathway curation, Pathway Commons will provide a mechanism and the technology for sharing. A key aspect of Pathway Commons is clear author attribution. Curation teams at existing databases must be supported by researchers to ensure they can keep performing their valuable work. Pathway Commons enables database providers to share their data in an efficient manner by avoiding duplication of effort and reducing software development overhead.
How is the data in Pathway Commons represented?
Pathway Commons uses the Biological Pathway Exchange (BioPAX) standard to represent data. Pathway databases that make their data available in this format can be imported into Pathway Commons. BioPAX is developed through a collaborative effort by many pathway databases.
What kind of biological concepts are represented?
Pathway Commons leverages the richness and versatility of BioPAX to store data. Details that can be included are biochemical reactions; gene regulatory networks; genetic interactions; proteins, small molecules, DNA, RNA, complexes and their cellular locations; complex assembly and transport; post-translational protein modifications; citations; experimental evidence; and links to other databases e.g. protein sequence annotation. Some information is only available in the downloaded BioPAX files.
How were the pathway data integrated?
Pathway data are downloaded directly from source databases. Each source has been created differently, some by manual extraction of pathway information from the literature and some by computational prediction. Warehouse data (canonical molecules, ontologies) are converted to BioPAX utility classes and saved as the initial BioPAX model, which forms the foundation for integrating data and for id-mapping. Pathway and binary interaction data (interactions, participants) are normalized next and merged into the database. Original reference molecules are replaced with the corresponding BioPAX warehouse objects.
We are a data provider. Why should we export our data to BioPAX?
Benefits of exporting your data to BioPAX and distributing it via Pathway Commons include:
- Your data will be used more: Through BioPAX and Pathway Commons, your data can reach more places, including many projects that rely on BioPAX for pathway data import and analysis. We pay attention to ensuring that you are clearly identified as the original data source so that you can receive credit. We log our website usage per data source and provide it back to you for your reporting needs.
- You will get more feedback and help with quality control: You can use the BioPAX validator to check your data against more than a hundred rules. We also automatically and manually check your exported data every release. Users of Pathway Commons often offer great feedback and whenever relevant we pass them back to you.
- Your data will be compatible with a range of software tools: There are more than 40 active tools that support BioPAX. Do you need web based visualization? You can use PCViz. Do you need graph and pattern searches? There are existing libraries for that. Do you want to use your data in Cytoscape or R? There are multiple apps that support BioPAX.
- We help you build your website and software tools: You will be able to automatically export your data to many other standard formats through BioPAX to e.g. SBGN, SBML, GSEA, SIF and linked data (RDF). Multiple software components are available to support more rapid application development, such as the powerful PaxTools Java library.
- Engage with a community of Pathway Informatics researchers: A key component of the BioPAX community is Pathway Data Providers like you. Through our online forums and face to face meetings, we were able to catalyze excellent convergence and interoperability between pathway databases and software tools. Comparing your data schema against others can give you excellent insights and an opportunity to introduce your ideas to other researchers.
- We will support your grant applications: Grant agencies often value support for open standard formats as evidenced by several previous grant evaluations. We will provide detailed support letters that explain your involvement and commitment to disseminate your data. We will also provide statistics of your data usage.
Contact
Post a question on the Pathway Commons help Google group.
Keep up-to-date by following the Pathway Commons announcements Google group.